Test dataset
This test simulation is intended to help in the development and testing of evaluation metrics. After running the short sequences through the aligner of your choice you can more quickly iterate your evaluation than on the larger data sets.
Root and Burnin
The root genome consisted of hg18 chr6:127,968,540-137,968,539 (10Mb) partitioned into two 5 Mb chromosomes with annotations populated from mgcGenes, knownGene, cpgIslandExt and ensGene tracks from the UCSC Table Browser. Details of infile set creation can be gleaned from the evolverInfileGeneration project
The root genome was evolved for a distance of 0.2 via 20 Evolver steps of 0.01, forming the simulation burnin. We used a much shorter initial burnin for this test simulation because this test set is intended to quickly iterate test evaluations and not make inferences. We wanted to make sure you could test evaluations using both the ancestor and root maf. The final genome of that burnin formed the ancestor for this simulation.
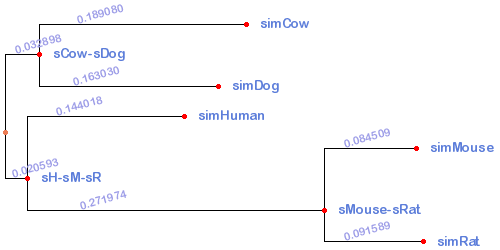
Tree
((simCow:0.18908,simDog:0.16303)sCow-sDog:0.032898,(simHuman:0.144018,(simMouse:0.084509,simRat:0.091589)sMouse-sRat:0.271974)sH-sM-sR:0.020593);
Summary Stats
| Genome | Chr | Size (bp) |
|---|---|---|
| simCow | chr0 | 5,531,984 |
| chr1 | 5,521,188 | |
| Total | 11,053,172 | |
| simDog | chr0 | 5,470,523 |
| chr1 | 5,518,726 | |
| Total | 10,989,249 | |
| simHuman | chr0 | 5,417,450 |
| chr1 | 5,507,808 | |
| Total | 10,925,258 | |
| simMouse | chr0 | 5,686,491 |
| chr1 | 5,785,878 | |
| Total | 11,472,369 | |
| simRat | chr0 | 5,639,451 |
| chr1 | 5,701,332 | |
| Total | 11,340,783 |
Files
Script to download and create the correct directory structure: downloadTest.sh
An analysis package has the following directory structure:
packageTest/
.. README.txt
.. annotations/
.. predictions/
.. sequences/
.. truths/
These directories may be populated with the following (expand all files):
- README.txt
- annotations/
-
version: 1
md5sum: 78e6dc3c21f842c38d2c8359e4962208
sha1sum: 90f58553a489e46095e415a992d31badf1e474e9
simTest.annots.gff.tar.gz (16 MB)
Annotations in gff format (optional).
version: 1
md5sum: a1df81a628c73e123cdcb1624bcccf64
sha1sum: 8a109c67e270d427595046ac2936e0d438fb1e9b
-
- predictions/ - place your .maf files in here. Below is an example set of three mafs to play with.
- simTest.examplePredictions.tar.gz (95 MB)
version: 1
md5sum: 30f05c4868a530d1aa38824574a09e9d
sha1sum: 1ecab02b20ad3815152fa5c7ec2bdc78f913bd7b
- simTest.examplePredictions.tar.gz (95 MB)
-
version: 1
md5sum: 579fbbfd2dfc8b07667eb0047d8a90da
sha1sum: b3da5a556626d4285698cedee0b71992e9234619
- Alignment to the MRCA, ancestor
simTest.ancestor.maf.tar.gz (37 MB)
aligns: {simMouse, simRat, sMouse-sRat, simHuman, sH-sM-sR, simCow, simDog, sCow-sDog, ancestor}version: 1
md5sum: 5c73438007aef3cd255403e5ad4f5483
sha1sum: 1cc74062d03de0cb556795bb13d83f84e6a9d72c
- Alignment to the Root, root
simTest.burnin.maf.tar.gz (48 MB)
aligns: {simMouse, simRat, sMouse-sRat, simHuman, sH-sM-sR, simCow, simDog, sCow-sDog, ancestor, root}version: 1
md5sum: 3fc9ff8328c3ed856a59a8235ccf0f5c
sha1sum: 16392bf7f360f45af220517a0bfcdb7318197beb
- Alignment to the MRCA and Root, with no paralogous blocks, ancestor.noparalogies, root.noparalogies
simTest.noparalogiyMafs.tar.gz (82 MB)
aligns: {simMouse, simRat, sMouse-sRat, simHuman, sH-sM-sR, simCow, simDog, sCow-sDog, ancestor, root}version: 1
md5sum: 28fcdd2181ca095b66a73d674d355ae9
sha1sum: 6dad4195c444ed59c1e0e19857a45266b56561c8
tree drawn using phyfi