Primate dataset
This simulation is intended to be a relatively simple alignment problem as there has been very little evolution that has taken place between the four lineages.
The root genome consisted of hg18 chr20, chr21 and chr22 with annotations populated from mgcGenes, knownGene, cpgIslandExt and ensGene tracks from the UCSC Table Browser. Details of infile set creation can be gleaned from the evolverInfileGeneration project
The root genome was evolved for a distance of 1.0 via 100 Evolver steps of 0.01, forming the simulation burnin. The final genome of that burnin formed the ancestor for this simulation.
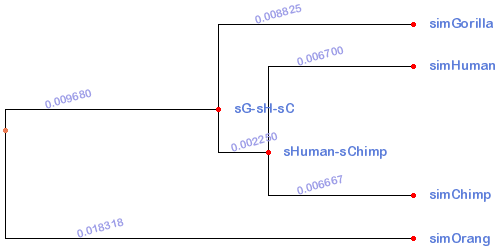
Tree
((simGorilla:0.008825,(simHuman:0.0067,simChimp:0.006667)sHuman-sChimp:0.00225)sG-sH-sC:0.00968,simOrang:0.018318);
Summary Stats
| Genome | Chr | Size (bp) |
|---|---|---|
| simChimp | A | 53,121,445 |
| B | 85,778,862 | |
| C | 35,661,804 | |
| D | 10,574,168 | |
| Total | 185,136,279 | |
| simGorilla | A | 53,120,926 |
| B | 85,848,133 | |
| C | 35,654,756 | |
| D | 10,570,608 | |
| Total | 185,194,423 | |
| simHuman | A | 53,106,993 |
| B | 85,835,872 | |
| C | 35,630,306 | |
| D | 10,572,275 | |
| Total | 185,145,446 | |
| simOrang | B | 85,903,762 |
| C | 35,683,973 | |
| D | 10,564,720 | |
| E | 37,692,687 | |
| F | 15,493,520 | |
| Total | 185,338,662 |
Files
Script to download and create the correct directory structure: downloadPrimates.sh
An analysis package has the following directory structure:
packagePrimates/
.. README.txt
.. annotations/
.. predictions/
.. regions/
.. sequences/
.. truths/
These directories may be populated with the following (expand all files):
- README.txt
- annotations/
simPrimates.annots.tar.gz (182 MB)
version: 1
md5sum: 7d337b5e4f7c6eeb8eeeda95c2c21271
sha1sum: 6be1156b635d81a29e63349831e003552c7a8277
simPrimates.annots.gff.tar.gz (162 MB)
Annotations in gff format (optional).
version: 2
md5sum: 9f4230f7c2349bd62abd6457be0a42c0
sha1sum: 095d141536819d9e0e8a55a60194d3252d75df55
- predictions/ - place your .maf files in here.
- regions/ - regional analysis takes place in here.
- sequences/
simPrimates.seqs.tar.gz (229 MB)
version: 1
md5sum: d817e8739c10a0ddfcbe37200545b7f9
sha1sum: a41f8d1aeb9b0afd58cb7805cc78f28416fdd72d
- truths/ - the true mafs are placed in here
- Alignment to the MRCA, ancestor
simPrimates.ancestor.maf.gz (143 MB)
aligns: {simHuman, simChimp, sHuman-sChimp, simGorilla, sG-sH-sC, simOrang, ancestor}version: 2
md5sum: 1e2417d2ae8b4cf2743d5e740b7c5ed3
sha1sum: fac50cbc86e759a1794208ee6e3f1b7d15e95fe2
- Alignment to the Root, root
simPrimates.burnin.maf.gz (581 MB)
aligns: {simHuman, simChimp, sHuman-sChimp, simGorilla, sG-sH-sC, simOrang, ancestor, root}version: 2
md5sum: 4fb72a9f14cf016c0d7b906d25e4731f
sha1sum: 6133a7de6788ed71fd2bfad9fea00fb9a439f163
- Alignments to the MRCA and Root, with no paralogous blocks, ancestor.noparalogies, burnin.noparalogies
simPrimates.noparalogyMafs.maf.gz (472 MB)
aligns: {simHuman, simChimp, sHuman-sChimp, simGorilla, sG-sH-sC, simOrang, ancestor, root}version: 2
md5sum: 3fc4fcb8fa64958f2a9d655b992387f7
sha1sum: c8714fd9adb64e27a615f08d35c900529034b2f7
- Alignment to the MRCA, ancestor
tree drawn using phyfi