Mammal dataset
The mammal dataset contains five species with length evolutionary distances between them. It is the most challenging simulated dataset in the Alignathon
The root genome consisted of hg18 (GRCh36) chr20, chr21 and chr22 with annotations populated from mgcGenes, knownGene, cpgIslandExt and ensGene tracks from the UCSC Table Browser. Details of infile set creation can be gleaned from the evolverInfileGeneration project
The root genome was evolved for a distance of 1.0 via 100 Evolver steps of 0.01, forming the simulation burnin. The final genome of that burnin formed the ancestor for this simulation.
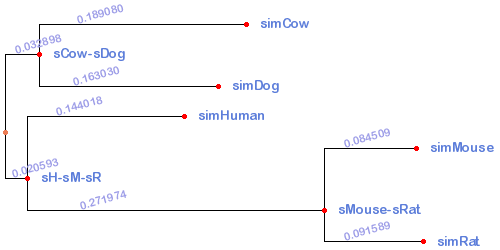
Tree
((simCow:0.18908,simDog:0.16303)sCow-sDog:0.032898,(simHuman:0.144018,(simMouse:0.084509,simRat:0.091589)sMouse-sRat:0.271974)sH-sM-sR:0.020593);
Sumamry Stats
| Genome | Chr | Size (bp) |
|---|---|---|
| simCow | A | 42,017,321 |
| B | 86,443,571 | |
| C | 33,408,597 | |
| D | 6,172,747 | |
| E | 24,983,699 | |
| Total | 193,025,935 | |
| simDog | A | 39,124,508 |
| D | 35,271,305 | |
| F | 64,906,724 | |
| G | 26,567,043 | |
| H | 20,782,131 | |
| I | 5,551,284 | |
| Total | 192,202,995 | |
| simHuman | D | 15,973,151 |
| F | 41,914,564 | |
| H | 2,880,482 | |
| I | 13,410,180 | |
| J | 88,398,963 | |
| K | 28,218,656 | |
| Total | 190,795,996 | |
| simMouse | A | 34,021,255 |
| F | 60,272,644 | |
| L | 71,158,916 | |
| M | 5,488,388 | |
| N | 16,897,397 | |
| O | 3,949,899 | |
| P | 7,132,917 | |
| Total | 198,921,416 | |
| simRat | A | 45,269,609 |
| O | 4,060,565 | |
| P | 7,089,915 | |
| Q | 54,146,922 | |
| R | 88,137,694 | |
| Total | 190,795,996 |
Files
Script to download and create the correct directory structure: downloadMammals.sh
An analysis package has the following directory structure:
packageMammals/
.. README.txt
.. annotations/
.. predictions/
.. regions/
.. sequences/
.. truths/
These directories may be populated with the following (expand all files):
- README.txt
- annotations/
simMammals.annots.tar.gz (203 MB)
version: 2
md5sum: bddd7ab44c51b45f79380f190fd7dfa0
sha1sum: 8f672d03b530ca9c3bff2729a50a90dc184aa82a
simMammals.annots.gff.tar.gz (196 MB)
Annotations in gff format (optional).
version: 2
md5sum: db3663a8ea555ffc274b4e7760d6b134
sha1sum: a9a66b6c684afaac5c5560170efa6583856c6ac3
- predictions/ - place your .maf files in here.
- regions/ - regional analysis takes place in here.
- sequences/
simMammals.seqs.tar.gz (303 MB)
version: 1
md5sum: a554a2151b3bbe269c2dcf6e07030ab7
sha1sum: eb7c21e112a8b049c9b69f20522f804a56c5cf01
- truths/ - the true mafs are placed in here
- Alignment to the MRCA, ancestor
simMammals.ancestor.maf.gz (652 MB)
aligns: {simMouse, simRat, sMouse-sRat, simHuman, sH-sM-sR, simCow, simDog, sCow-sDog, ancestor}version: 2
md5sum: 4bab2832a972a26a9a43af150096295e
sha1sum: 31395977ecbe948cc0bb4455a5db618e9f543fd2
- Alignment to the Root, root
simMammals.burnin.maf.gz (1.2 GB)
aligns: {simMouse, simRat, sMouse-sRat, simHuman, sH-sM-sR, simCow, simDog, sCow-sDog, ancestor, root}version: 2
md5sum: 0a4c595644a806e7342ec3be62893f39
sha1sum: 8bfa54e894a5f276bdfda385e8eeddec90c718bb
- Alignments to the MRCA and Root, with no paralogous blocks, ancestor.noparalogies, burnin.noparalogies
simMammals.noparalogyMafs.maf.gz (1.4 GB)
aligns: {simMouse, simRat, sMouse-sRat, simHuman, sH-sM-sR, simCow, simDog, sCow-sDog, ancestor, root}version: 2
md5sum: 28fcdd2181ca095b66a73d674d355ae9
sha1sum: 6dad4195c444ed59c1e0e19857a45266b56561c8
- Alignment to the MRCA, ancestor
tree drawn using phyfi