Fly dataset
The fly data set is comprised of 20 real fly genomes in variou states of completion, from the nearly-complete Drosophila melanogaster dm3 (chromosome sequences) to the fragmentary D. rhopaloa droRho (34,000 contigs).
These genomes are real and therefore do not have a root genome or a burnin process.
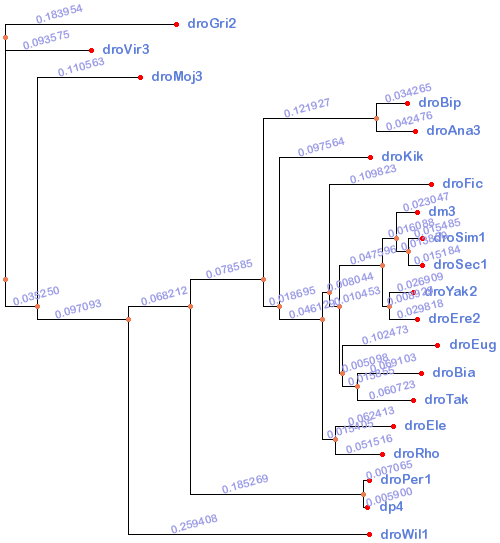
Tree
This is a combination of the phylogeny presented in the modENCODE white paper proposing the sequencing of eight additional fly genomes (genome.gov) courtesy of Artyom Kopp (UC Davis) and the phylogeny used by UCSC for the 15-way inserct alignment. The Kopp tree lacked droSim1 and droSec1 which were added by normalizing the branch lengths between the dm3 branches on the two trees. Extraneous species were trimmed using tree_doctor from PHAST. This tree is provided for progressive aligners that need a guide tree and will be used in the analysis for StatSigMa-w.
((droGri2:0.183954, droVir3:0.093575):0.000000, (droMoj3:0.110563, ((((droBip:0.034265, droAna3:0.042476):0.121927, (droKik:0.097564, ((droFic:0.109823, (((dm3:0.023047, (droSim1:0.015485, droSec1:0.015184):0.013850):0.016088, (droYak2:0.026909, droEre2:0.029818):0.008929):0.047596, (droEug:0.102473, (droBia:0.069103, droTak:0.060723):0.015855):0.005098):0.010453):0.008044, (droEle:0.062413, droRho:0.051516):0.015405):0.046129):0.018695):0.078585, (droPer1:0.007065, dp4:0.005900):0.185269):0.068212, droWil1:0.259408):0.097093):0.035250);
Summary Stats
| Genome | Chr | Size (bp) |
|---|---|---|
| dm3 | 2L | 23,011,544 |
| 2LHet | 368,872 | |
| 2R | 21,146,708 | |
| 2RHet | 3,288,761 | |
| 3L | 24,543,557 | |
| 3LHet | 2,555,491 | |
| 3R | 27,905,053 | |
| 3RHet | 2,517,507 | |
| 4 | 1,351,857 | |
| M | 19,517 | |
| U | 10,049,037 | |
| Uextra | 29,004,656 | |
| X | 22,422,827 | |
| XHet | 204,112 | |
| YHet | 347,038 | |
| Total | 168,736,537 |
Files
Readme with notes on the build process to download and create the flies.seq.tar.gz file (not necessary to download, data is contained in downloadFlies.sh below, this link is here for the sake of a paper trail,): createFlies.txt
Script to download and create the correct directory structure: downloadFlies.sh
An analysis package has the following directory structure:
packagePrimates/
.. README.txt
.. annotations/
.. predictions/
.. regions/
.. sequences/
.. truths/
These directories may be populated with the following (expand all files):
- README.txt
- annotations/ - at the moment there are no suggested annotations for the fly package.
- predictions/ - place your .maf files in here.
- regions/ - regional analysis takes place in here.
- sequences/
version: 2
md5sum: ffbd16a9979bf3d233f59b4422de3315
sha1sum: 14575f2df189ac03c6d56dcb5b9ce14f2bafa7ba
- truths/ - leave this empty
tree drawn using phyfi